Microbiomes in sympatric populations
Identifying environmental and host-specific drivers of bacterial symbiont composition in natural populations of sympatric Daphniidae
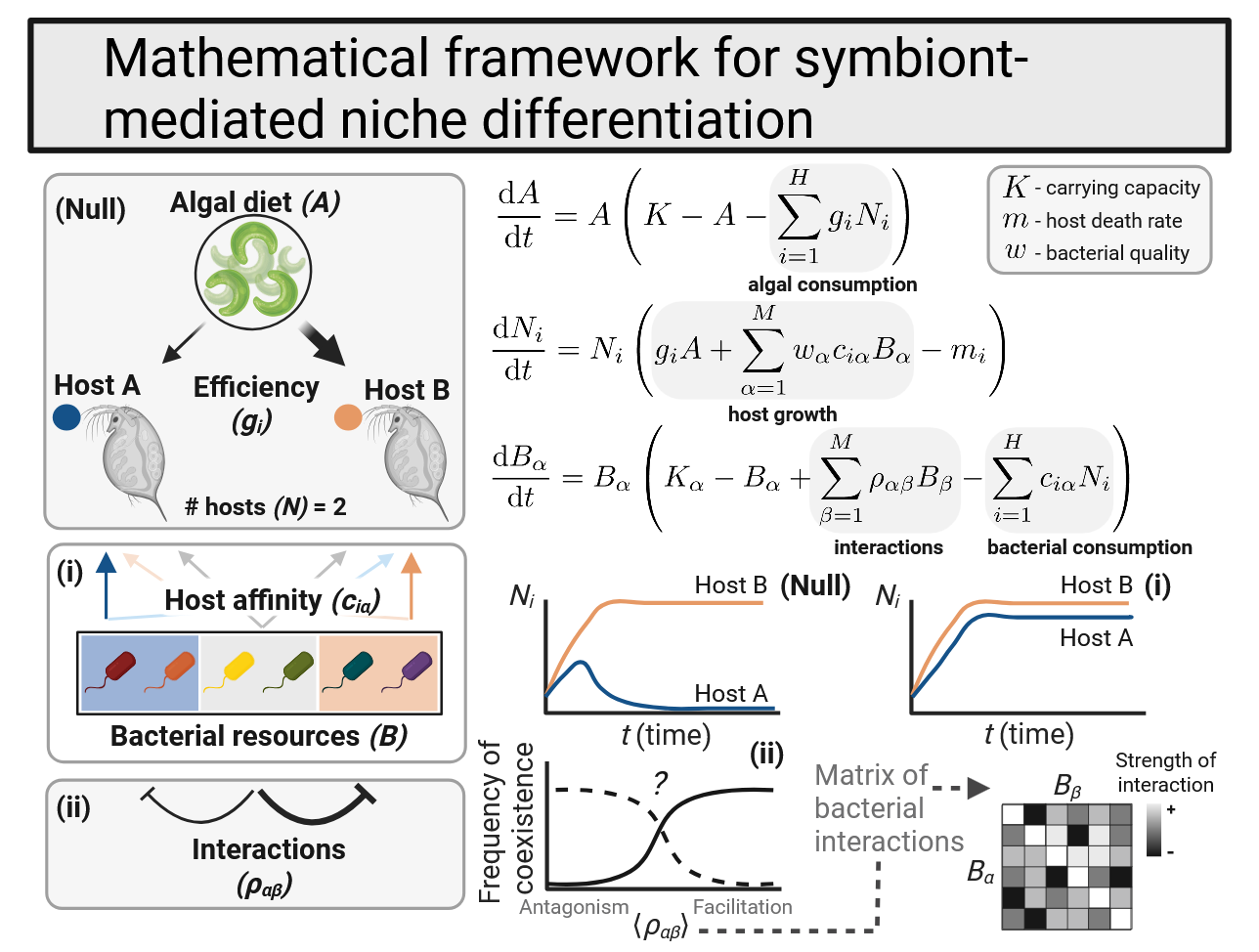
Ecologists have long considered the factors enabling the species coexistence. To use an oversimplified example, niche theory would expect that two organisms using one resource would necessarily competitively exclude each other, with the organism capable of more efficient resource-use persisting in the environment. The so-called “paradox of the plankton” is that an amazing array of biodiversity exists in-spite of this simple expectation: among, for example, phytoplankton that seemingly share very similar resource-use patterns and ecology. Explanations for coexistence in phytoplankton communities and other systems with high biodiversity and functional redundancy have led to numerous scientific papers, with hypotheses ranging from temporal differences in resource-use, fine-scale resource partitioning, and food web dynamics sustaining complexity (just to name a few!). I am interested in the ways microbial resources might be partitioned to additionally explain the coexistence of ecologically similar species. A comprehensive understanding of the ecological interactions sustaining biodiversity is key to preserving it in light of global change, which threatens the stability of long-standing community dynamics.
In collaboration with the Duffy lab at the University of Michigan I have been analyzing sequencing data collected from a series of lakes in southeastern Michigan. These data represent the microbiome of multiple types of daphnids, important species in lake ecosystems where they graze down phytoplankton, serve as a food source for numerous fish species, and aid in nutrient cycling. Daphnia have long served as a model for toxicology, epidemiology, and increasingly eco-evolutionary study of host-microbe interactions. Dr. Duffy’s group has carved out a niche in bringing together field surveys of host-parasite interactions and controlled laboratory studies to tackle fundamental questions in epidemiology. I am particularly interested in sympatric species of daphnids in these lakes, and determining if there is any partitioning of environmental bacterial communities that might hint at niche differentiation in the wild. The lab’s datasets also contain rich metadata describing parasite burden, adding a layer of complexity that I hope to dive into further.
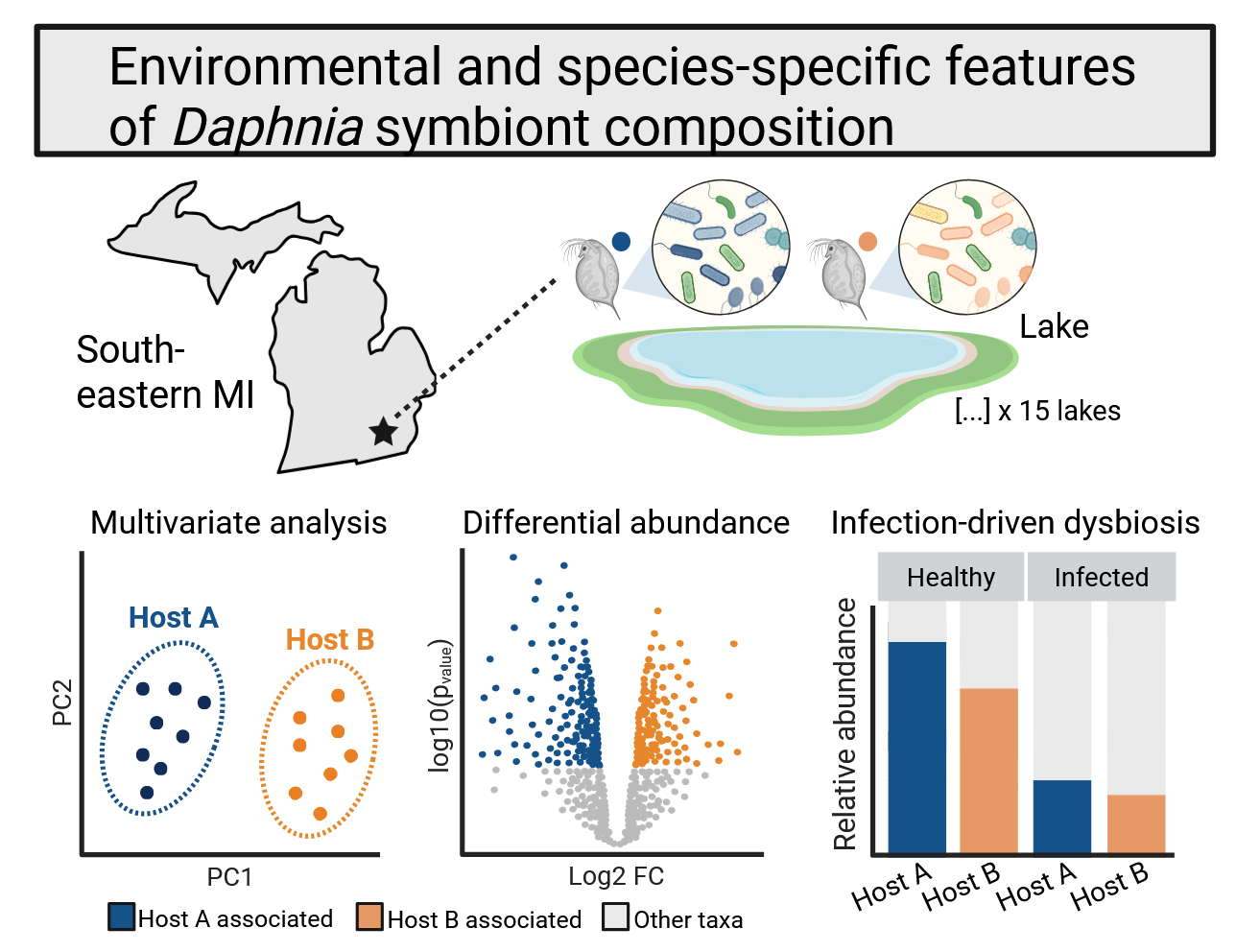
In the longer term, I am interested in the broader phenomenon of niche differentiation as mediated by host-associated microbial communities. Several systems have reported differences among sympatric species’ microbiota and alluded to gut-associated bacterial communities as an axis of niche separation. Can we glean general rules about levels of complexity in host communities and features of the biotic environment that would facilitate host coexistence?
Associated references:
Arellano A.A.*, Cooper R.O.*, Hite J.L., Dziuba M.K., Cressler C.E., Duffy M.A. Environmental and species-specific features of microbiome assembly in wild daphnids. (In prep)
* authors contributed equally